He fetal genetic diagnosis is fundamental in prenatal careand recent progress Prenatal Tri-Exome Sequencing (i.e., simultaneous sequencing of the coding regions of the fetal, maternal, and paternal genomes) proposes clinical benefit. However, due to the invasive nature of embryo sampling, its use is limited to embryos with identifiable structural abnormalities. This limitation prevents many monogenic disorders from being diagnosed because they do not manifest in the prenatal period, resulting in a large number of newborns falling ill. fatal childhood disorder And seriously debilitating.
Development Non-invasive fetal testing using circulating DNA Maternal blood sampling has revolutionized prenatal testing, but its application is primarily limited chromosome mutation Due to the low resolution of existing tests.
To address these limitations, they developed a non-invasive prenatal screening using tri-exome deep sequencing. This comprehensive method has high sensitivity for detecting Fetal single nucleotide variants, small insertions and deletions, high copy number mutations and chromosomal aneuploidies from circulating DNA obtained from maternal blood samples. (Figure 1). This proof-of-concept approach leverages error-corrected ultra-deep trio-exome sequencing to detect fetal variants. afresh (new, non-inherited mutations) with high precision. These are the main reasons severe child development disorders,
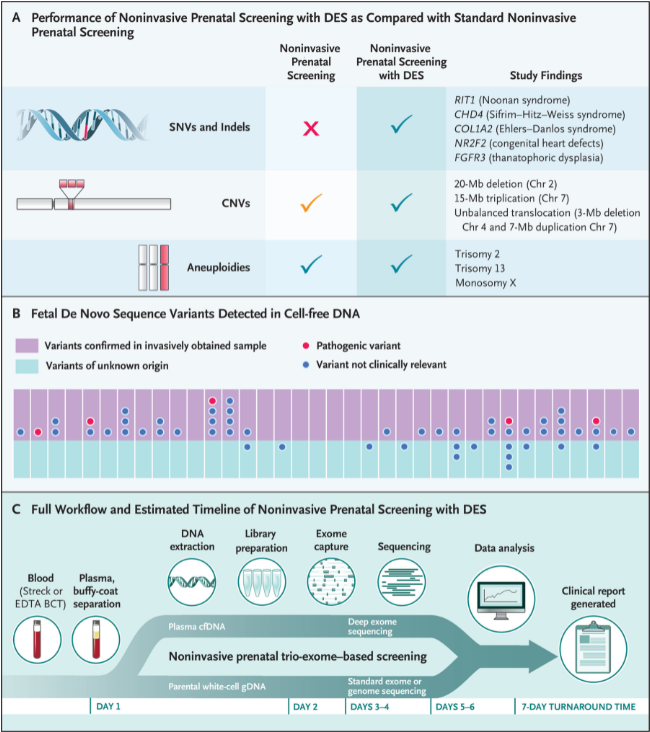
Figure 1: Performance and workflow of non-invasive prenatal screening with tri-exome deep sequencing.
In this study they included 36 pregnant women and their partners. The main inclusion criteria were measured nuchal translucent At least one fetal anomaly, at least 5 mm, was identified during the first or second trimester of pregnancy or both. they made a Ultra-deep exome sequencing in free DNA Extracted from maternal plasma, providing high sequencing coverage per sample. A custom process was used to accurately name and filter variants, which included error correction using unique molecular identifiers, site-specific noise modeling, and embryo fraction estimates ranging from 3.72–19%.
Tri-exome analysis including maternal plasma and matched parental blood samples was performed to identify new fetal and paternally inherited variants. The performance of non-invasive prenatal screening using three-exome deep sequencing was evaluated using the results obtained from this new method in comparison to invasive whole-exome sequencing, whole-genome sequencing, and analysis. microarray For all cases, chromosomes from chorionic villi or amniotic fluid samples.
The following were identified Embryonic Pathogenic Variants afreshThose considered clinically relevant in pregnancy: Four single nucleotide mutations, one small deletion, two autosomal trisomies, one sex chromosome aneuploidy, two high copy number variants, and one unbalanced translocation., All mutations detected by this non-invasive method were confirmed in embryo samples obtained invasively. These were aligned with the embryo’s phenotype and provided a genetic diagnosis In 11 out of 36 pregnancies. No additional variants were identified by invasive analyzes in 36 cases. Therefore, complete agreement has been observed between non-invasive and invasive prenatal analysis, with 100% detection of pathogenic variants. afresh,
To assess the representation of the fetal exome in circulating DNA, they expanded the analysis to include all paternally inherited fetal sequence variants within the coding region. The average sensitivity of the new method for detecting these mutations in all protein-coding genes was 95.12%.
In a subset of plasma samples, this new technology detected very few variants of unknown origin that were not identified in the corresponding invasively obtained samples. However, none of these were considered clinically relevant or required further testing.
In this non-invasive method, parental samples were included, which allows carrier screening to identify carriers in both parents. Pregnancies at higher risk for recessive disorders, This testing approach demonstrates that it can accurately determine fetal inheritance of a paternal mutation in the case when the mother is not a carrier. Invasive testing will be required to confirm the presence of the maternally inherited variant in the fetus.
This type of prenatal screening, non-invasively and comprehensively, provides a means of detecting a wide range of genetic disorders Without posing any risk to the fetus or mother. Integrating this technology into routine prenatal care along with ultrasound screening will provide an opportunity to improve early detection rates, reduce the number of invasive procedures, and facilitate rapid intervention. Although larger studies are needed to test this hypothesis, the findings of this investigation (Figure 1B) would predict a low false positive rate. All positive results should be confirmed by prenatal invasive diagnostic testing. Non-invasive prenatal testing using tri-exome deep sequencing provides timely results, with a turnaround time of approximately 7 days (Figure 1C).
The findings support further investigation of deep exome sequencing of circulating DNA to detect pathogenic variants in fetuses with and without detectable structural abnormalities.